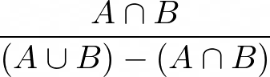true positives (TP) matched correctly,
false positives (FP) incorrectly segmented objects and
false negatives (FN) incorrectly missed objects.
precision = TP / (TP + FP),
recall = TP / (TP + FN) and
F1 = 2 x precision x recall / (precision + recall)
true positives (TP) matched correctly,
false positives (FP) incorrectly segmented objects and
false negatives (FN) incorrectly missed objects.
precision = TP / (TP + FP),
recall = TP / (TP + FN) and
F1 = 2 x precision x recall / (precision + recall)
This function removes out of focus blur from the source images using neural networks. It is intended for widefield images and works best for thick samples. It is a preferred choice for under-sampled images, whereas deconvolution is a preferred method for well-sampled images.
See Deconvolution.
Clarify.ai requires valid image metadata (similar to deconvolution). It is a parameterless method which does not increase the resolution and does not denoise the image however it can be combined with NIS.ai > Denoise.ai. Check the Denoise.ai check box next to a channel to perform denoising first before clarifying. Check this check box only for very noisy images with SNR value smaller than 20.
To handle the out-of-focus planes correctly, it is important to know how exactly the image sequence has been acquired. Select the proper microscopic modality from the combo box.
Depending on the Modality setting, set the pinhole/slit size value and choose the proper units.
Specify magnification of the objective used to capture the image sequence.
Enter the numerical aperture of the objective.
Enter the refraction index of the immersion medium used. Predefined refraction indexes of different media can be selected from the pull-down menu.
Enter the image calibration in μm/px.
Check if you need to create new document. Otherwise the clarifying is applied to the original image.
Select which channels will be clarified and which will be denoised. You can also adjust the emission wavelength. To revert the changes, click .
If checked, the clarifying preview is shown in the original image.
Confirms the settings and performs the clarifying.
Closes the window without executing any process.
(requires: Local Option)(requires: 2D Deconvolution)(requires: 3D Deconvolution)
Opens the Restore.ai dialog window. This function is designed to be used when denoise and deconvolution processes are combined. It can be applied on all types of fluorescence images (widefield, confocal, 2D/3D, etc.).
To handle the out-of-focus planes correctly, it is important to know how exactly the image sequence has been acquired. Select the proper microscopic modality from the combo box.
Specify magnification of the objective used to capture the image sequence.
Enter the numerical aperture of the objective.
Enter refraction index of the immersion medium used. There are some predefined refraction indexes of different media in the nearby pull down menu.
Enter the image calibration in μm/px.
Image channels produced by your camera are listed within this table. You can decide which channel(s) shall be processed by checking the check boxes next to the channel names. The emission wavelength value may be edited (except the Live De-Blur method).
Note
Brightfield channels are omitted automatically.
Performs image denoising with the use of neural networks. This function is used especially for static scenes because moving objects may get blurred.
Detects whether cells are present in the brightfield image or not. Works even for out of focus images. Node outputs:
Verdict is 1 if cells are present, 0 if they are not present.
Confidence of the detection, ranging from 0 (not confident) to 1 (very confident).
Detects cells and outputs a binary image with dots on the detected cell centers.
Works only on images with bright field modality and in a narrow Z range around focus (+/- 25 µm).
For the function description please see Enhance.ai.
Selects the trained network from a file (click to locate the *.eai file).
Opens metadata associated with training of the currently selected neural network.
For the function description please see Convert.ai.
Selects the trained network from a file (click to locate the *.cai file).
Opens metadata associated with training of the currently selected neural network.
For the function description please see Segment.ai.
Selects the trained network from a file (click to locate the *.sai file).
Opens metadata associated with training of the currently selected neural network.
Reveals post-processing tools and restrictions used for enhancing the results of the neural network.
For the function description please see Segment Objects.ai.
Selects the trained network from a file (click to locate the *.oai file).
Opens metadata associated with training of the currently selected neural network.
Reveals post-processing tools and restrictions used for enhancing the results of the neural network.
Uses Cells Localization.ai to segment the image into areas with cells and into homogeneous areas without cells. The resulting binary image is equal to 0 at the cells and to 1 at the homogeneous areas. The segmentation fails if less than 10 cells are found.
Select the channel for segmentation.
Check to invert the resulting binary (change 0s to 1s and vice versa).
Selects an appropriate trained AI file according to the sample objective magnification. Output is expected to be used as a dynamic input parameter for another AI node. Paths to the trained AI files, either relative or absolute, can be defined using a standard regular expression.
(requires: Local Option)
Estimates the Signal to Noise Ratio (“SNR”) value as it is used with Autosignal.ai (AutoSignal.ai).
Calculates the average precision to evaluate AI on objects. This node has two inputs - GT (Ground Truth) and Pred (Prediction). It compares the ground truth binary layer (A) and predicted binary layer (B) generated by segmentation using AI. It also pairs the objects from both layers and classifies them (based on the IoU threshold) into:
Based on these numbers it calculates:
Defines a threshold above which two overlapping objects are considered correctly matched - threshold of quantity:
.
Calculates the average precision to evaluate AI on objects. This node has two inputs - GT (Ground Truth) and Pred (Prediction). It compares the ground truth binary layer (A) and predicted binary layer (B) generated by segmentation using AI. It also pairs the objects from both layers and classifies them (based on the IoU threshold) into:
Based on these numbers it calculates:
Defines a threshold above which two overlapping objects are considered correctly matched - threshold of quantity:
.
(requires: NIS.ai)
 NIS.ai
NIS.ai