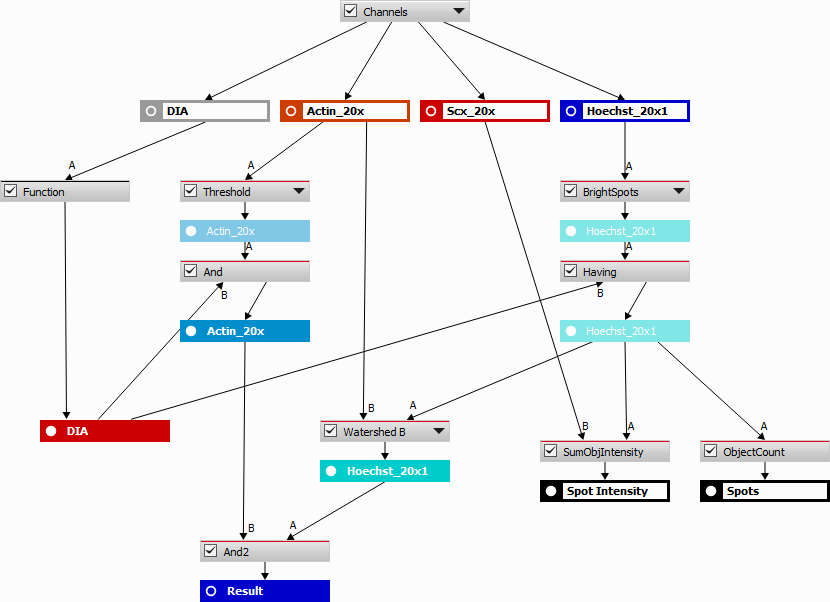
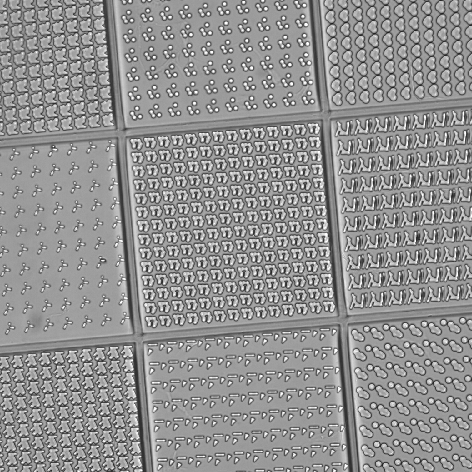
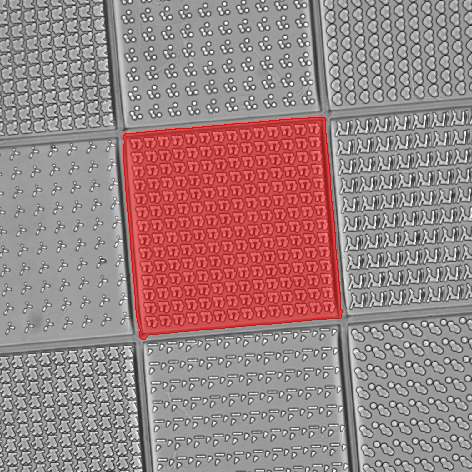
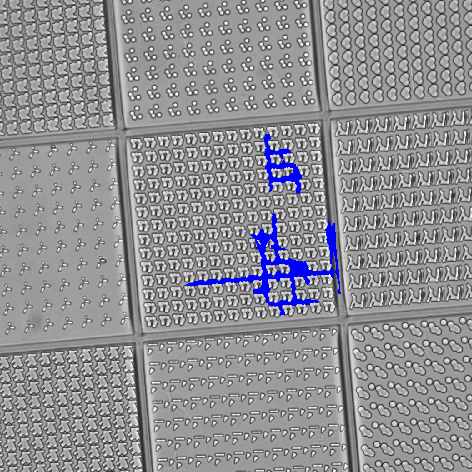
In this example, TopoChip is used as a sample to screen the bio-active surface topography. The goal is to count the number of cells (“Actin_20x” channel) and their nuclei (“Hoechst_20x1” channel) inside the central square (TopoUnit) and to measure the intensity (signal) of the “Scx_20x” channel for each cell.
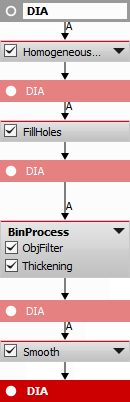
At first, the “DIA” channel is segmented using Homogeneous Area. Holes are filled and filtering, thickening and smooth is applied to correctly detect the central square topography unit on which the measurement will be performed. Using a camera ROI would not bring satisfactory results because of the slight tilt of the sample.
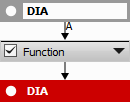
The whole sequence is then grouped into a single “Function”.
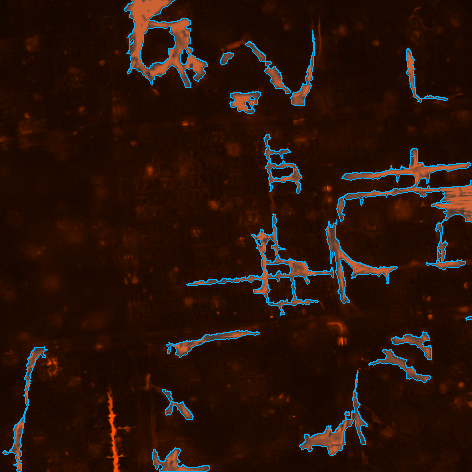
To detect cells, “Actin_20x” channel is thresholded. Because the area of interest is just the central square unit, logical conjunction (And node) is done between the result and the segmented “DIA” image.
To detect the nuclei in the central TopoUnit, bright spots are detected on the “Hoechst_20x1” channel and binary objects from the segmented “DIA” binary result being inside or touching the bright spots are found (Having node).
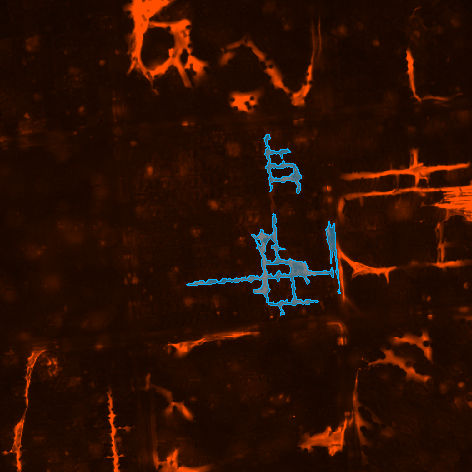
Assuming each cells has one nucleus, watershed algorithm is applied to the result of the Having node and another logical And is done between the watershed result and “Actin_20x” result to properly assign nuclei to the respective cells.
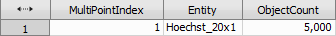
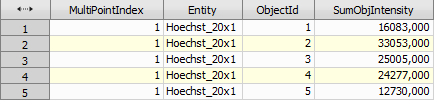
Finally, object count calculates the number of cells (bright spots) present in the central TopoUnit and Sum Object Intensity calculates the amount of “Scx_20x” signal per each cell.