(requires: CA FRET)
This “how to” expects you to perform a ratio experiment.
See also: [Ca 2+] Ion Concentration Measurement.
The intended procedure consists of the following portions:
Setting up Optical Configurations
6D: Time and MultiChannel Set Up
Create/ Save/ Open ROIs
Time Measurements Set Up
Capturing data set
Calcium Calibration
Export to Excel/ Text File
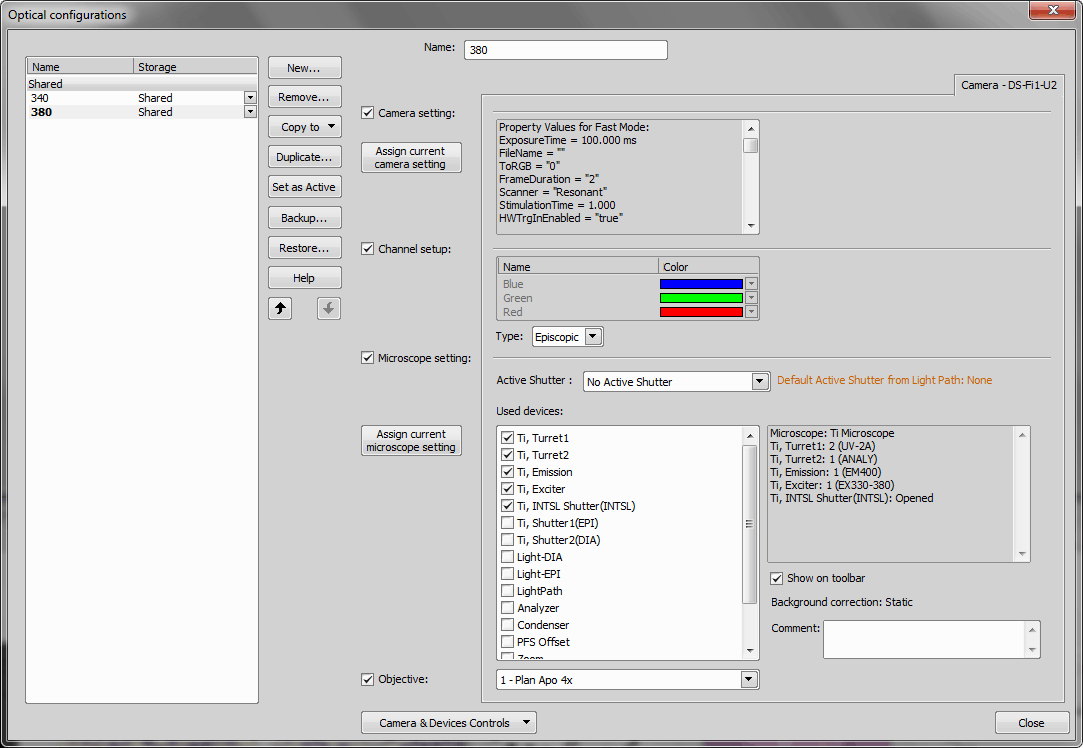
Create two optical configurations containing filter settings (use the  Calibration > New Optical Configuration
Calibration > New Optical Configuration  command). Call them “340” and “380” (indicating excitation wavelengths).
command). Call them “340” and “380” (indicating excitation wavelengths).
Note
If different camera settings (exposure, EM gain, etc) are desired for individual channels, then create microscope + camera settings optical configurations.
Active Shutter will most likely be the EPI Shutter.
The only difference between these two Optical Configurations may be the filter state of the excitation of 340 and excitation of 380.
Run the  Applications > 6D > Define/Run ND Acquisition
Applications > 6D > Define/Run ND Acquisition  command.
command.
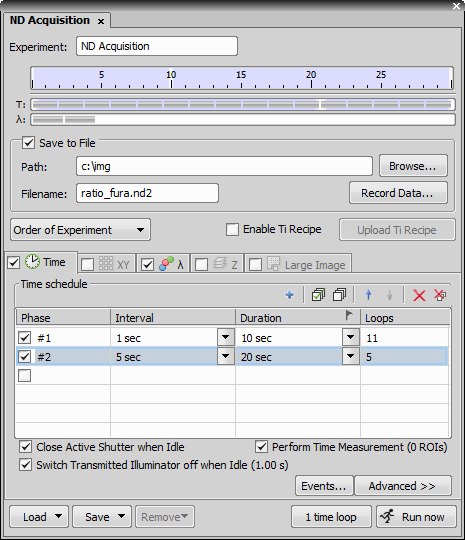
In the Time tab, adjust the settings according to the image:
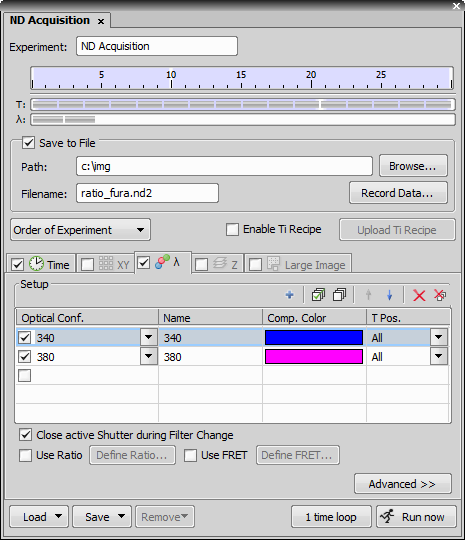
In the Lambda (wavelength) tab, define two channels:
In the case of 340 and 380, this is a dual excitation experiment. These two channels will share the same emission (510nm). In this case, the component color (pseudocolor) of each of these channels will be the same because NIS-Elements uses the emission wavelength as the default to set the component color. If it is desired to have differing component colors for 340 and 380, then select the drop down list of component colors to change (or both) of the lambdas.
Note
At this point, it is recommended (if not completed already) to optimize the correct camera exposures for 340 and 380. In a typical Fura-2 experiment, 380 emission will appear dimmer than 340, an indicator that cellular calcium is bound, not free. When establishing optimal exposure, be cognizant that in an experiment that induces a release of calcium, the emission of 380 will increase. Set exposures to match the dynamic range of the camera and also the full length of the time-lapse and possible biological responses to drug additions.
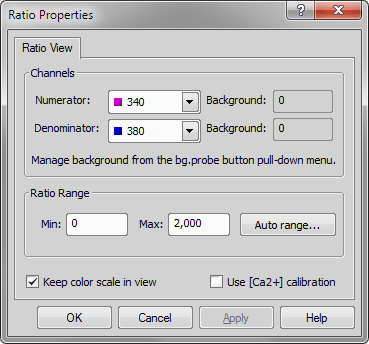
In the Time tab, select Use Ratio and press the button.
The Ratio Range is a parameter that specifies the Intensity Modulation Display (IMD). Using the option will automatically scale the IMD based on ratios in the dataset. (Auto range may not be available at the onset of an experiment/ Timelapse because the data for the ratio has not yet been generated.)
Note
Use [Ca2+] calibration will be an important item to check at later time after acquisition. There are instances when [Ca2+] calibration can be specified before acquisition. Values from a previously run experiment can be inserted here.
Confirm that Perform Time Measurement is checked within the Time tab.
Open up Time Measurement dialog by the
 View > Analysis Controls > Time Measurement
View > Analysis Controls > Time Measurement  command.
command.
Press the button in the 6D dialog window.
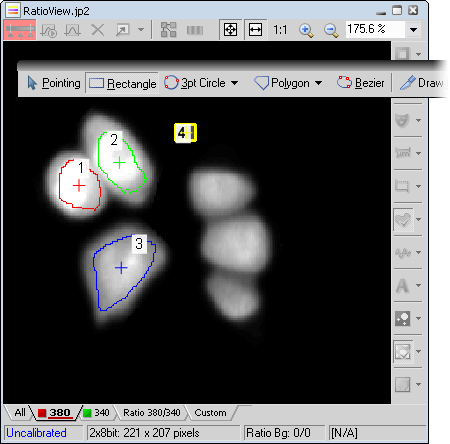
In the ROI editor, select the Polygon tool.
Draw several ROIs and one designated to indicate background.
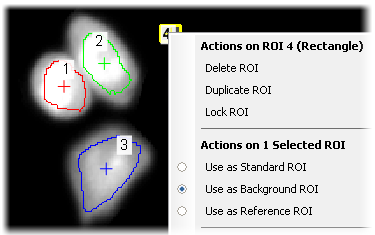
Right-click the background ROI (#4) and select Use as Background ROI from the context menu.
Once a background ROI is specified, it immediately begins to perform a subtraction and it will be noticeable in the IMD display. The background probe icon
 on the image window will also become active. This drop-down menu provides more options for how to utilize this background ROI. In this case, “Keep Updating Background Offset” is selected.
on the image window will also become active. This drop-down menu provides more options for how to utilize this background ROI. In this case, “Keep Updating Background Offset” is selected.Save ROIs to a file to be able to load it later. Right-click the ROI button
 on the right image toolbar and select Save ROI As. The ROIs will be saved in a common image format (TIFF, BMP, PNG).
on the right image toolbar and select Save ROI As. The ROIs will be saved in a common image format (TIFF, BMP, PNG).
Open up Time Measurement dialog by the
 View > Analysis Controls > Time Measurement
View > Analysis Controls > Time Measurement  command (if it has not been already open).
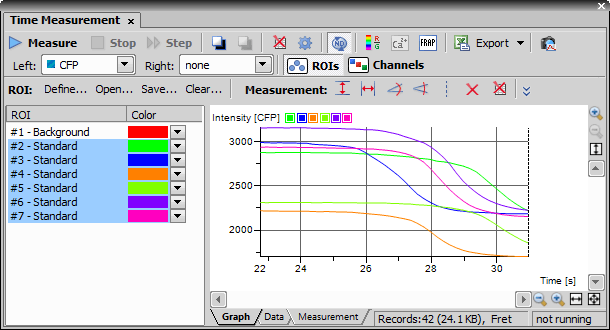
command (if it has not been already open).Highlight (select) all ROIs in order to be displayed in the graph.
At this point in the experiment set up, notice what will be graphed during the Timelapse. Define Left and Right axes display.
Select the display mode - ROIs mode vs. Channels mode. Graphing
 ROIs will graph each cell/ROI individually over time. Graphing
ROIs will graph each cell/ROI individually over time. Graphing  Channels will graph the average intensity/ ratio of each cell/ ROI over time.
Channels will graph the average intensity/ ratio of each cell/ ROI over time.
Note
These are graphing options for the during of the Timelapse, but can be modified after acquisition. Choices made at this point in the experimental set up do not affect the data values or limit graph choices post acquisition.
In the 6D dialog window, press the Run Now button. A new image will be captured.
You will be able to observe ratio graphs during the acquisition. This is critical to understand if there is a change in response to the solutions added to the specimen to create ratio responses - a change in the ratio of bound and free calcium in the cells. The live graph is an indication of whether or not there was a response (e.g. if the dose was toxic). This is basically a tool to allow the researcher how to plan the course of the experiment.
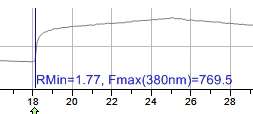
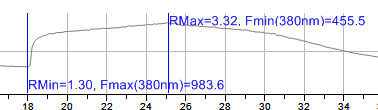
Once the full dataset has been measured through the time measurements dialog, it is now possible to set the calcium calibration- normalizing the data off of the experimental Rmin and Rmax.
Select the
 icon.
icon.Pick Rmin value from graph - This is also the Fmax (maximum fluorescence intensity of 380. (Generally speaking this is after EGTA is added and near the start of the calibration routine of the ratio experiment before ionomycin is added).
Pick Rmax value from graph - This is also the Fmin (minimum fluorescence intensity of 380. (Generally speaking this is after ionomycin is added- highest levels of free calcium).
Click .
What becomes available after calcium calibration
Dataset has a new channel tab named [Ca2+] with corresponding IMD display.
Graph options for Left and Right now include Ca+2 as one of the drop down options.
In the Time Measurement window, select the Data to Excel option from the Export pull-down menu.
Press the button.
See Exporting Results.
Note
If MS Excel is not installed on your computer, the only way to export the data is Data to File and Graph to Raster File / Graph to Clipboard